Bigger and better RNA scaffolds for organising proteins
Researchers from Aarhus University and Caltech have developed a method to build much larger, though still nanosize, RNA scaffolds than previously thought possible using RNA origami. The method is based on new software that has been made available online so other researchers can use it to develop biosensors, nanorobots and medicine – including vaccines, for example.
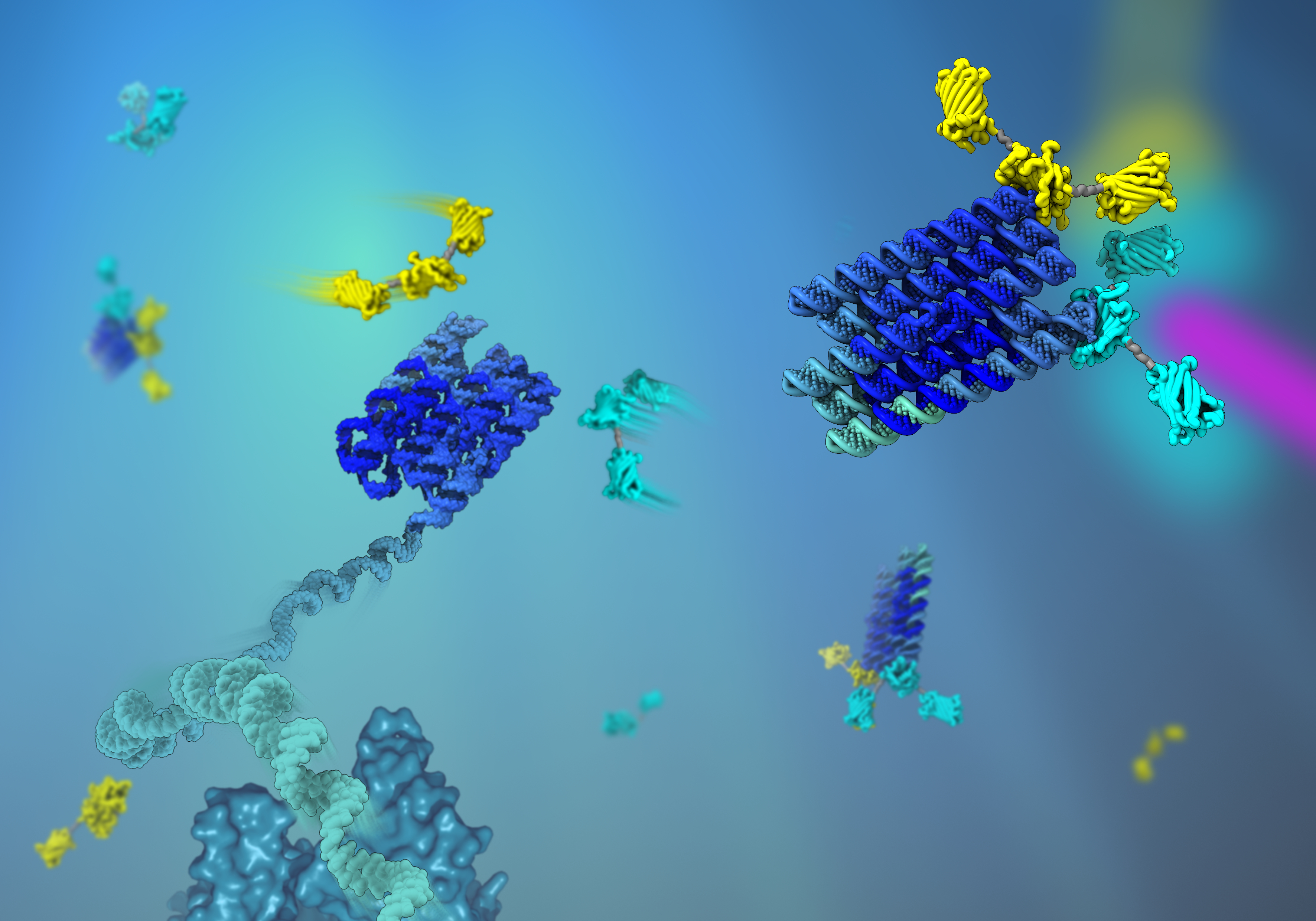
You normally encounter scaffolding when it is used to support structures and temporary work platforms in connection with construction work. But your own cells use scaffolding too, just in nanosize and made from proteins, to organise cellular processes and to construct your body. These protein scaffolds have inspired bioengineers to design “artificial” nano scaffolds that can be used to organise biological processes, for example to optimise the biological production of valuable chemicals.
In a new study published in Nature Chemistry, researchers from Aarhus University and the California Institute of Technology take a major step forward in the rational design of RNA scaffolds for organising proteins. By using the RNA origami method, they have produced much larger scaffolds than previously thought possible.
The design software that made the study possible has been made available online by the researchers, including tutorials and a dedicated server, so that other researchers can build RNA scaffolds for their own purposes.
Origami with limitations
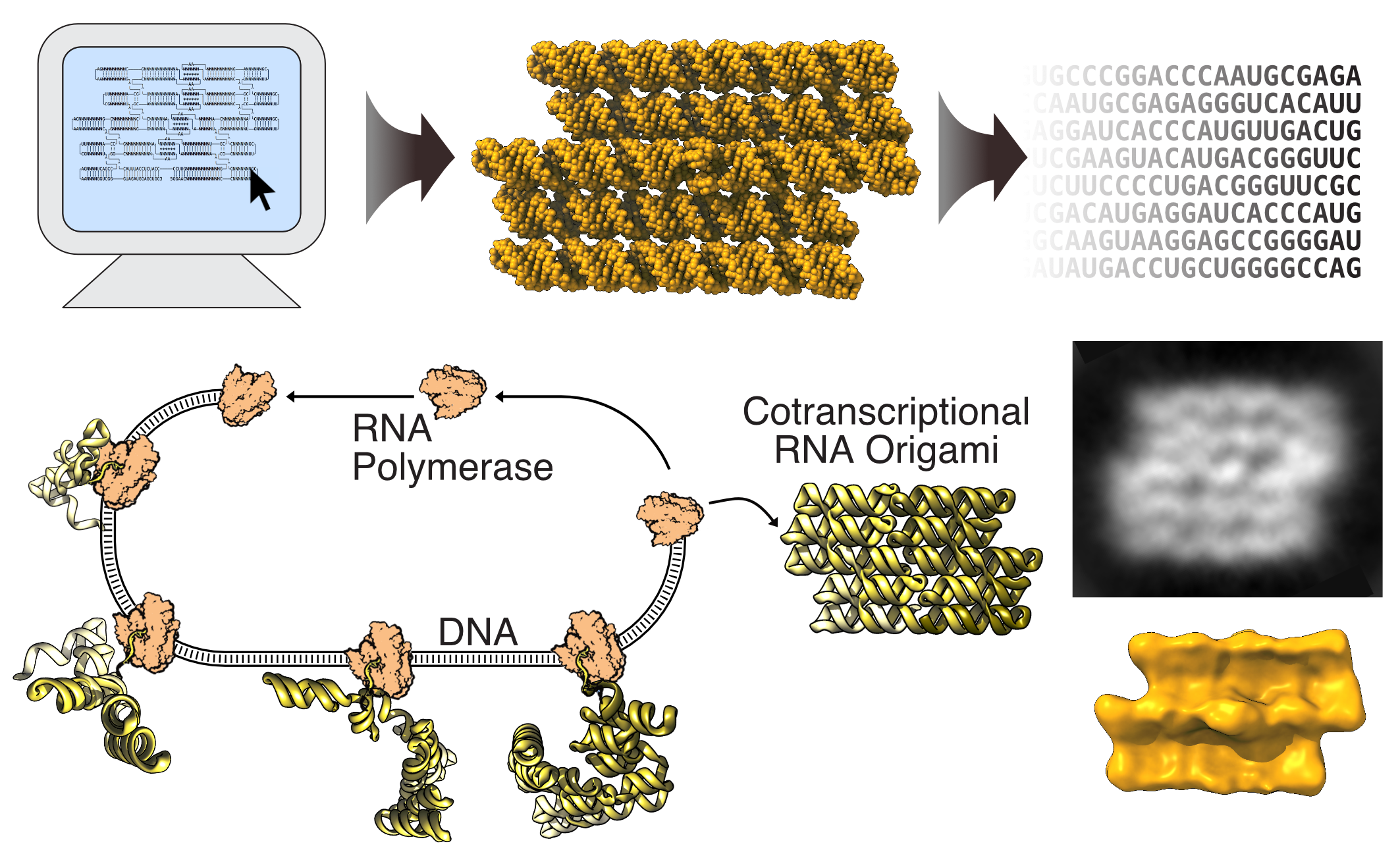
The RNA origami method was introduced in 2014 as a set of RNA elements that could be combined like Lego bricks to construct RNA nanostructures that can fold from a single strand. A important finding was that these RNA structures could be folded during transcription by an RNA polymerase, which is a desired property that makes it possible to genetically encode and express RNA nanostructures in cells.
However, the RNA origami method had serious limitations:
"In the original paper we reached a size of 450 nucleotides in length for folded RNA origami with a rather low yield. We also reached the limits of the RNA design software available at that time. The current study addresses these limitations," explains Ebbe Sloth Andersen, associate professor at Aarhus University.
Bigger and better RNA scaffolds
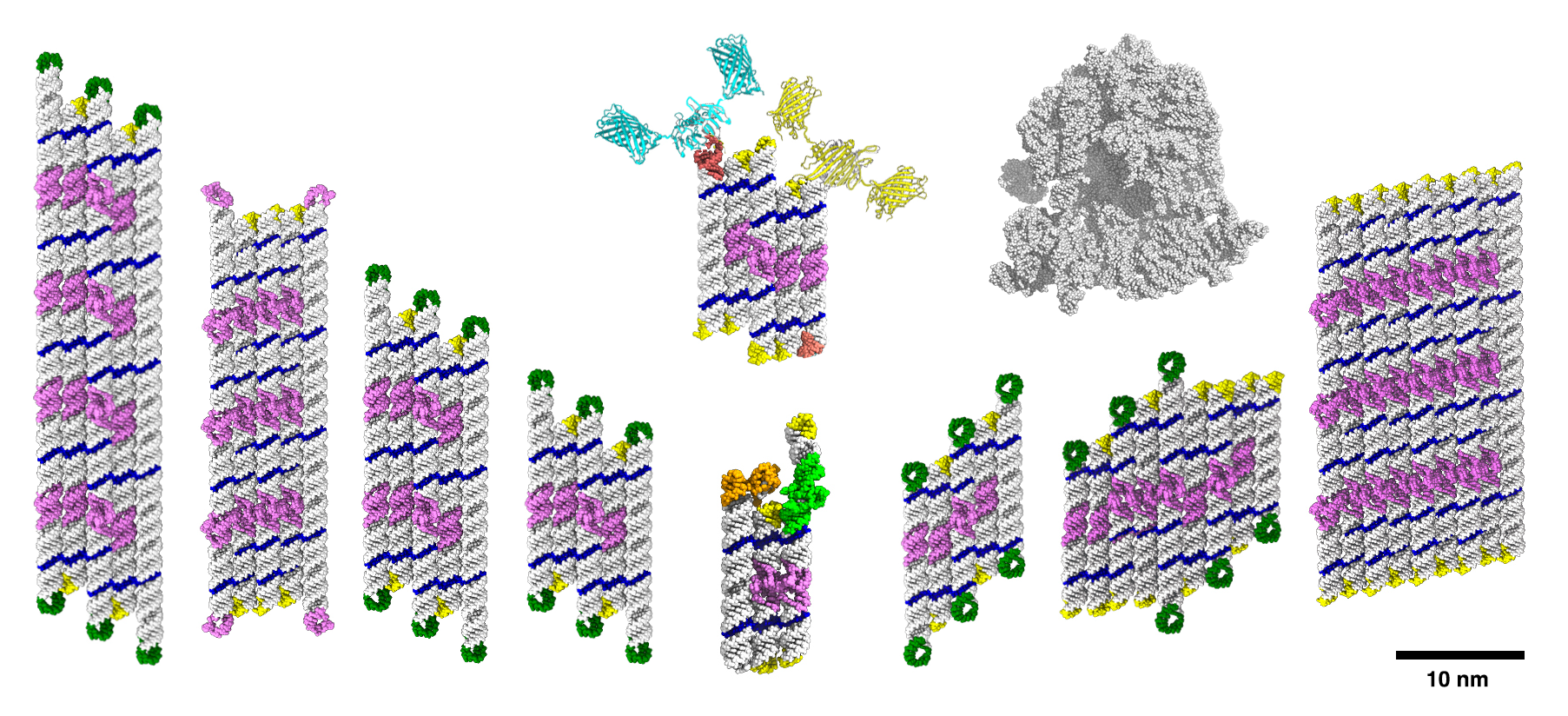
The current study was made possible by the development of software that can design a large panel of RNA origami structures. RNA origami structures are designed as flat sheets of RNA helices that have now been expanded by either the number of stacked helices or along the helix axis. The study found that RNA origami structures can be scaled in both directions up to approx. 2,000 nucleotides in length with up to 24 long-range interactions.
"We were surprised to produce well-defined RNA structures of this size. Previously we had only a limited set of eight long-range interactions to pick from, and a major breakthrough was finding a way to produce new sequences for these programmable connectors,” says Cody Geary, assistant professor at Aarhus University, who developed the project both at Caltech and Aarhus University.
The research team noticed that the folding yield dropped substantially for the largest structures. To understand why, they conducted an experiment to investigate effects of different folding paths. By using the same structure with different folding paths and investigating the yield of folded structures by electron microscopy, postdoc Ewan McRae observed folding defects in RNA origami structures.
They then used the RNA origami structures with the best folding yield for scaffolding experiments.
Never wrote software before
The software package that made the study possible automates the main steps in the RNA origami design process. The package is composed of a library of scripts that were all written by Cody Geary, who had never written a software package.
"But perhaps it was an advantage. In the final analysis, the program runs almost totally backwards to convention, solving the most difficult constraints at the very end. I think that’s essentially the secret to what makes it work so well,” says Geary.
The design of RNA origami structures starts with the construction of a blueprint composed of predefined modules.
- The first program, called RNAbuild, translates the blueprint into atomic models which allow the user to gradually build an RNA origami structure and get feedback on the overall shape.
- The second program, called RNApath, estimates the order of folding and topological barriers, which help the user evaluate different choices of folding path.
- The third program, called Revolvr, designs an RNA sequence that will fold into the blueprint structure. Revolvr solves many of the previous limitations of RNA design algorithms by being able to design structures of large size and with multiple long-range interactions using relatively short calculation times.
In minutes, the software is able to provide results that used to take days and weeks using other existing software. Software that, in theory, should be able to design such structures but can’t.
Once the sequence is designed, a DNA template is ordered from a gene synthesis company. With the DNA template in hand, you can make the RNA structure using a simple RNA transcription. In order to see whether the RNA structures have formed as expected, advanced microscopy methods are used to visualise the structure.
Animation of the folding of an RNA origami, where orange and red colors indicate potential topological problems for the folding process. Animation: Cody Geary Future applications of RNA-origami
The current study was conducted with pure components in a test tube, but RNA origami can in principle be expressed in cells. Studies are beginning to emerge where RNA origami structures are genetically encoded and expressed in cells or used as medical nanoparticles that can regulate blood clotting.
"I imagine that we will be able to design increasingly complex structures much more freely in three dimensions and even be able to engineer moving parts within these structures. This type of progress will lead to the development of more compact designs that can function as nanomachines or nanorobots," says Cody Geary.
According to Ebbe Sloth Andersen, the future looks bright for the use of RNA nanostructures in both medicines and synthetic biology.? With mRNA vaccines delivered in lipid nanoparticles, the prospect of delivering RNA medical particles is becoming a reality, and genetically expressed RNA origami has many possible applications within synthetic biology.
"We are now working on using RNA scaffolds as tools in biotechnology. For this purpose, we are developing RNA origami as biosensors to detect metabolites in cells, as scaffolds for regulating gene expression, and as scaffolds for enzymes to direct metabolic production," says Ebbe Sloth Andersen.
Additional informations | |
| We strive to ensure that all our articles live up to the Danish universities' principles for good research communication(scroll down to find the English version on the web-site). Because of this the article will be supplemented with the following information: | |
| Funding | The studies have received financial support from the European Research Council (ERC), the National Science Foundation (NSF), the Office of Naval Research (ONR), the Carlsberg Research Foundation, the Independent Research Fund Denmark, Natural Sciences and Engineering Research Council of Canada. |
| Read more | Link to the article in Nature Chemistry Tutorial for RNA design software and webserver are available at: https://bion.au.dk/software/rnao-design/ |
| Kontakt | Associate professor Ebbe Sloth Andersen. Email:esa@inano.au.dk Mobile:+45 41178619 Assistant professor Cody Geary. Email: geary@inano.au.dk Professor Paul Rothemund. Email:pwkr@dna.caltech.edu. Phone:+1 626-390-0438 |
